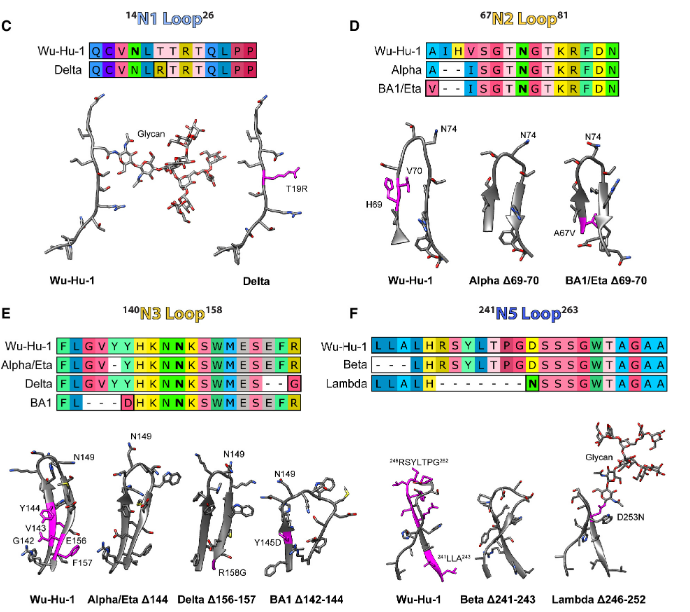
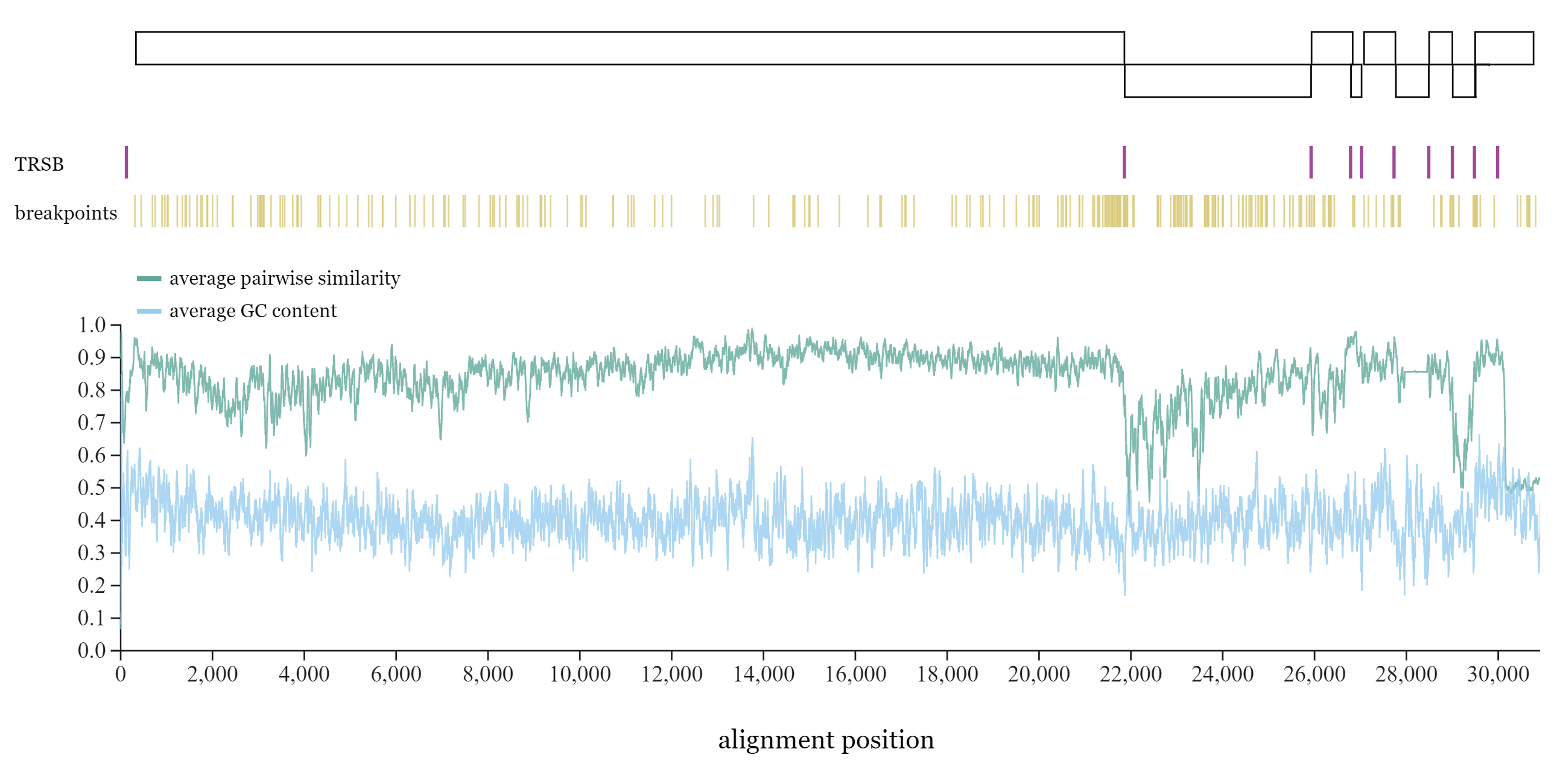
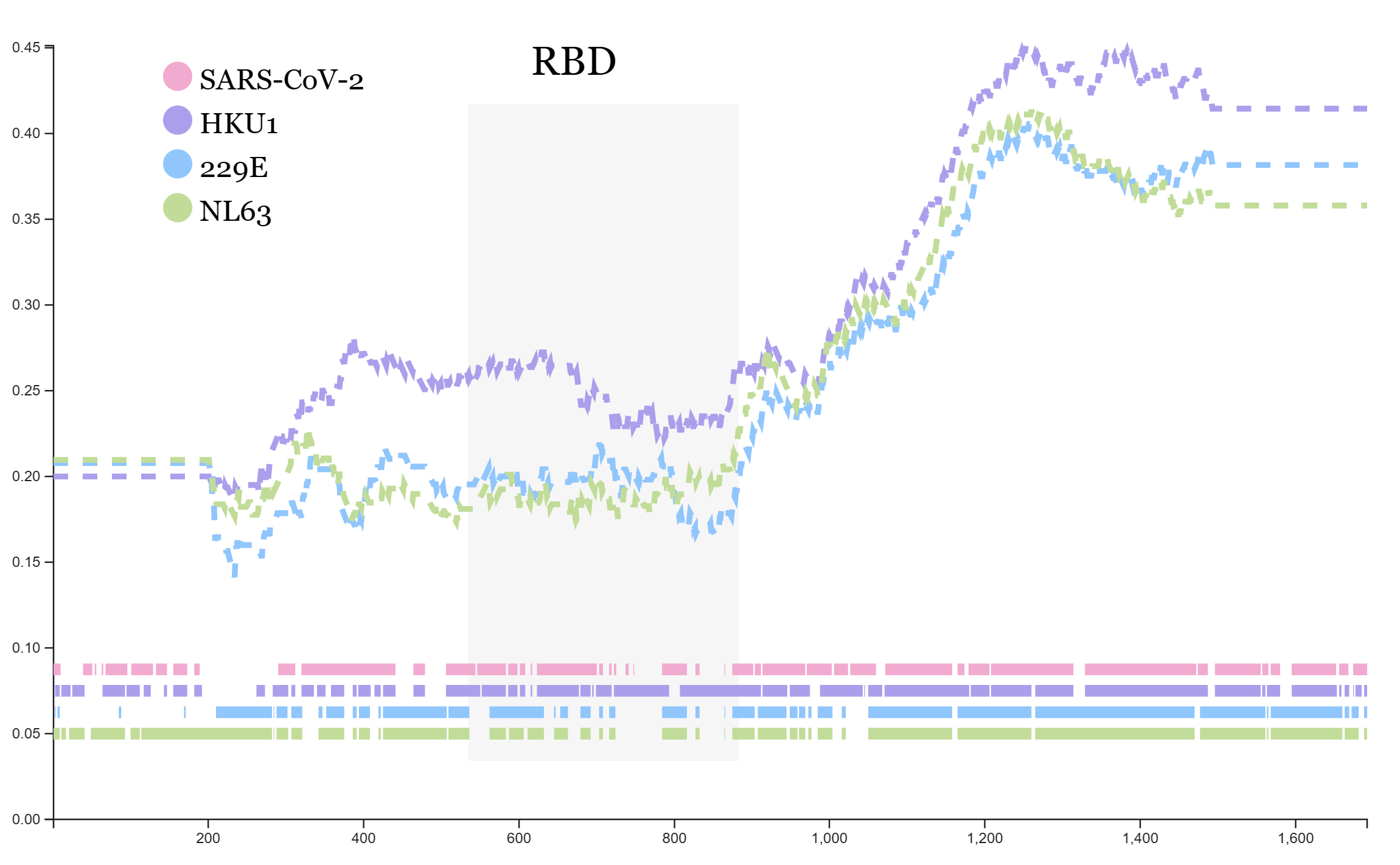
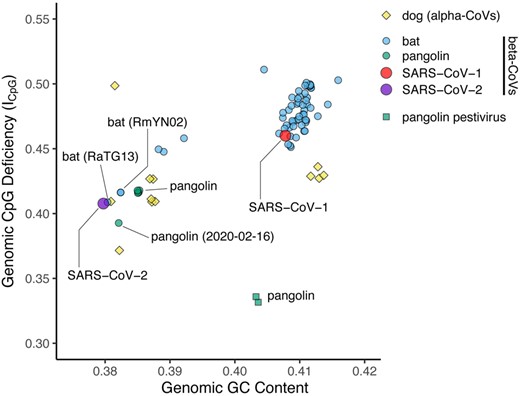
Public Engagement
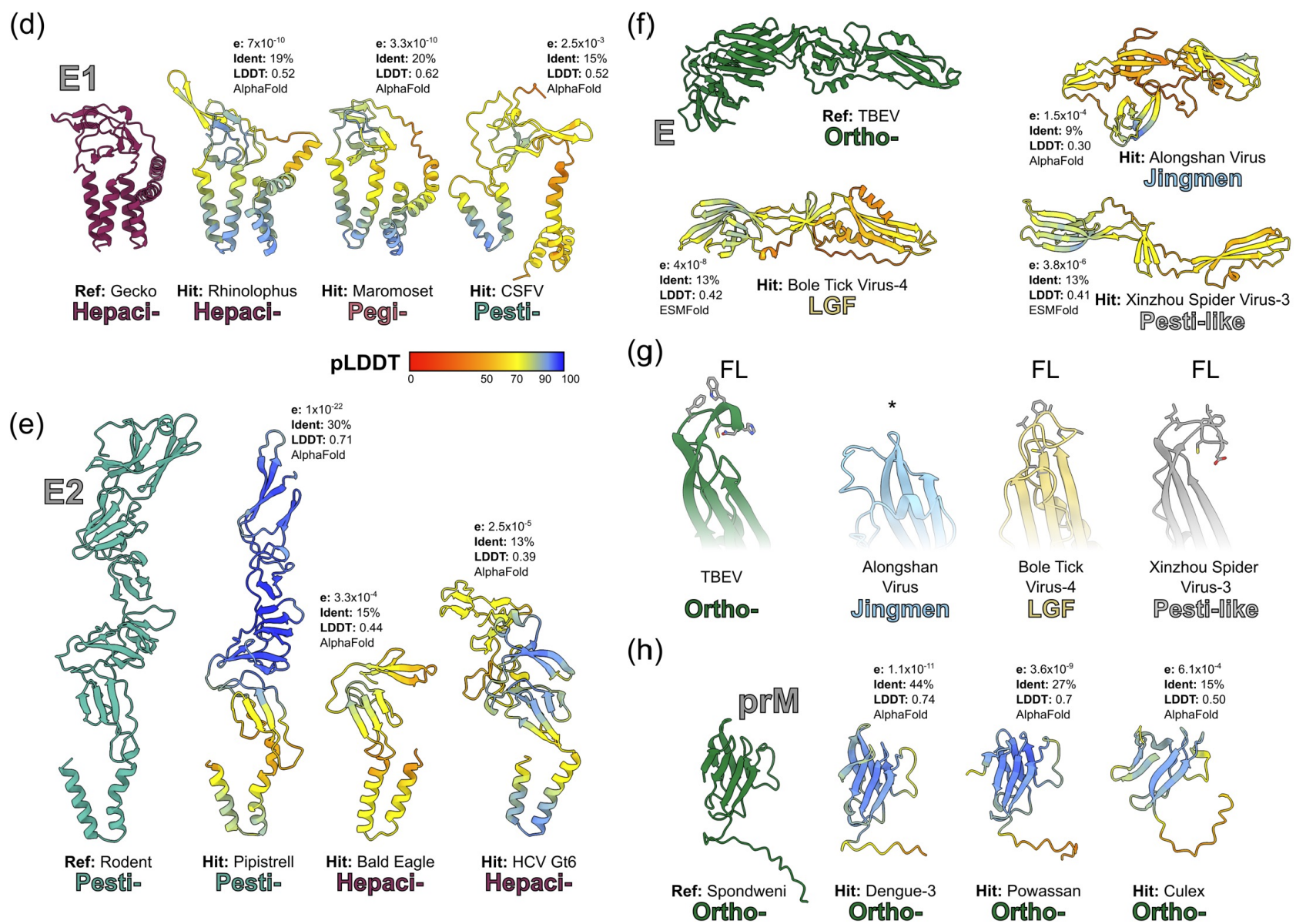
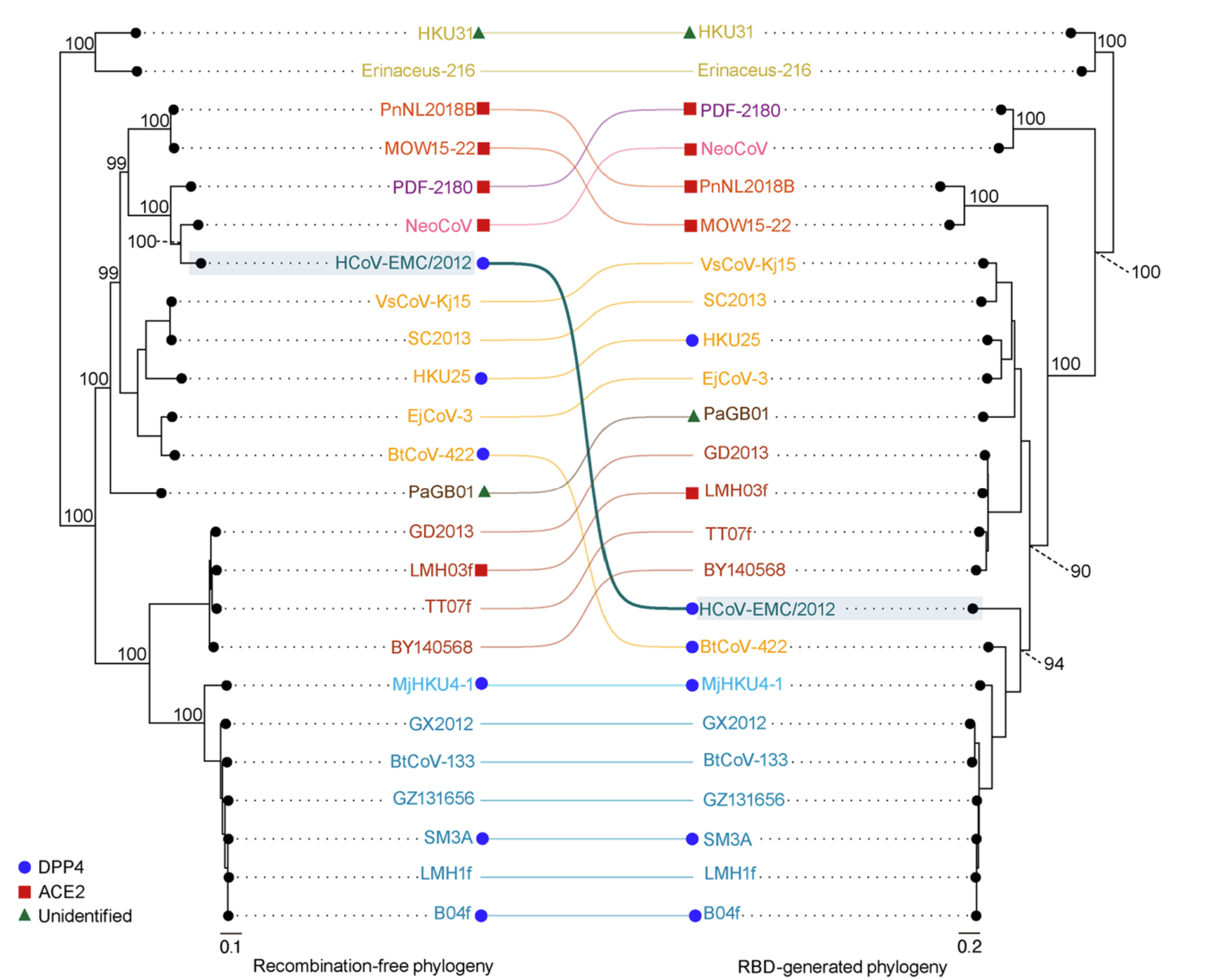
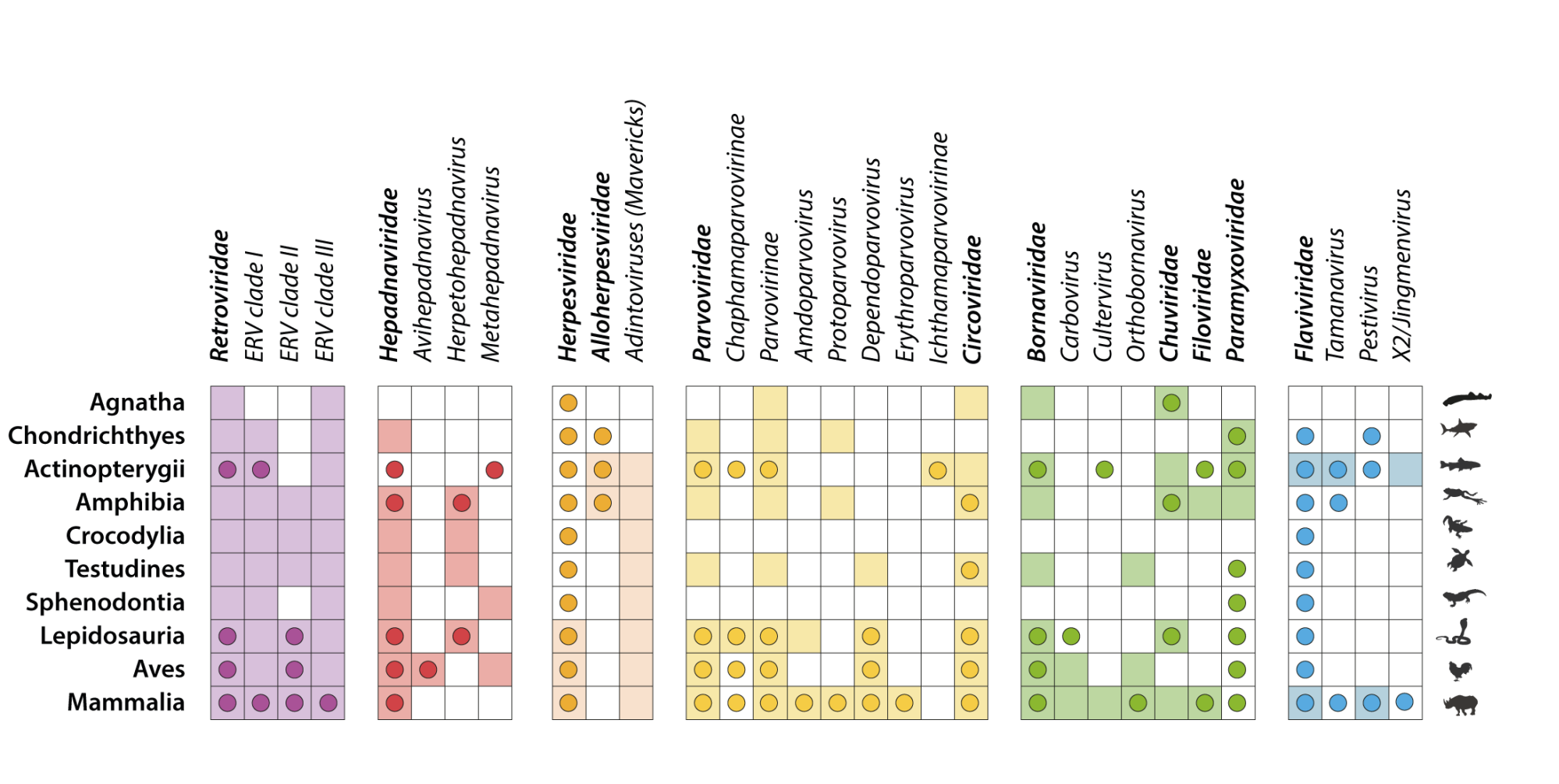
Exploring the molecular interplay between viruses and their reservoir hosts' immune systems can aid our understanding of how these viruses can potentially spill over into new hosts. Have a watch of the clip below where I explain our work on resurrecting an ancient bat defence against SARS-related coronaviruses
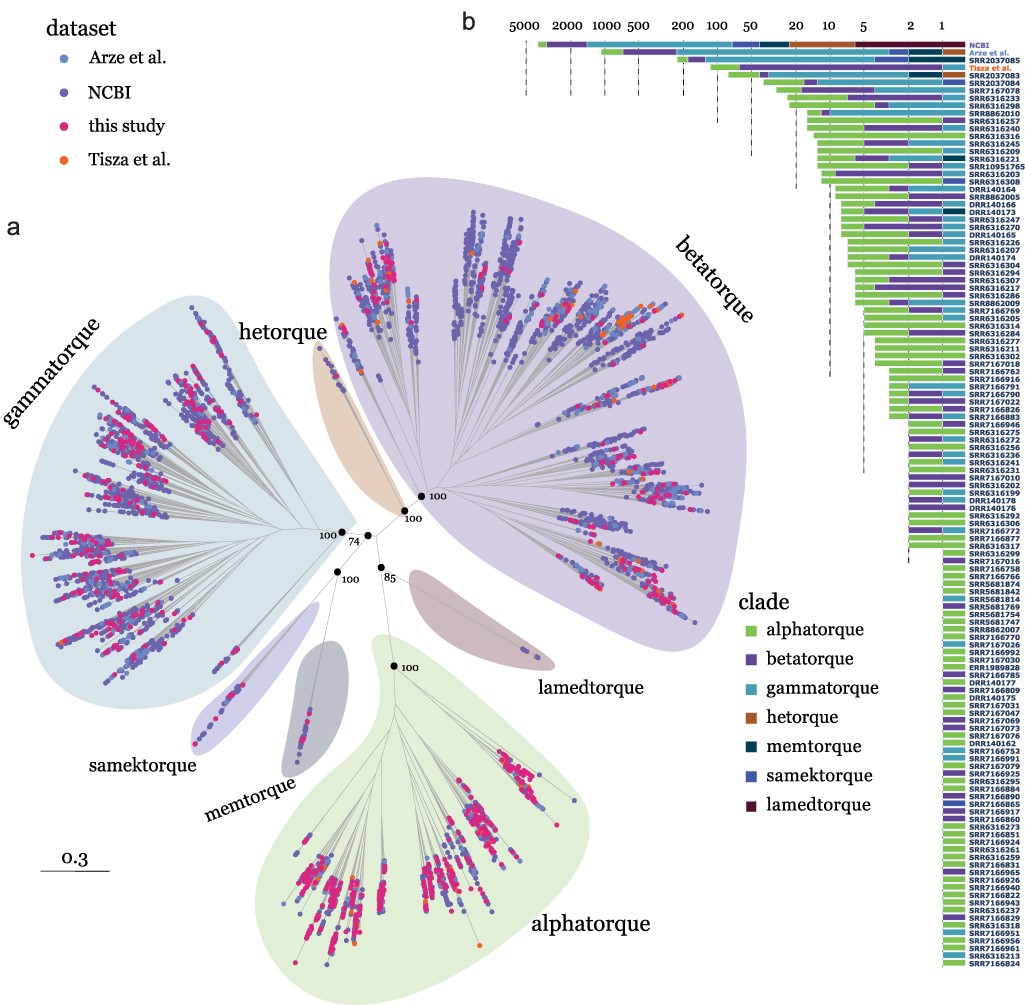
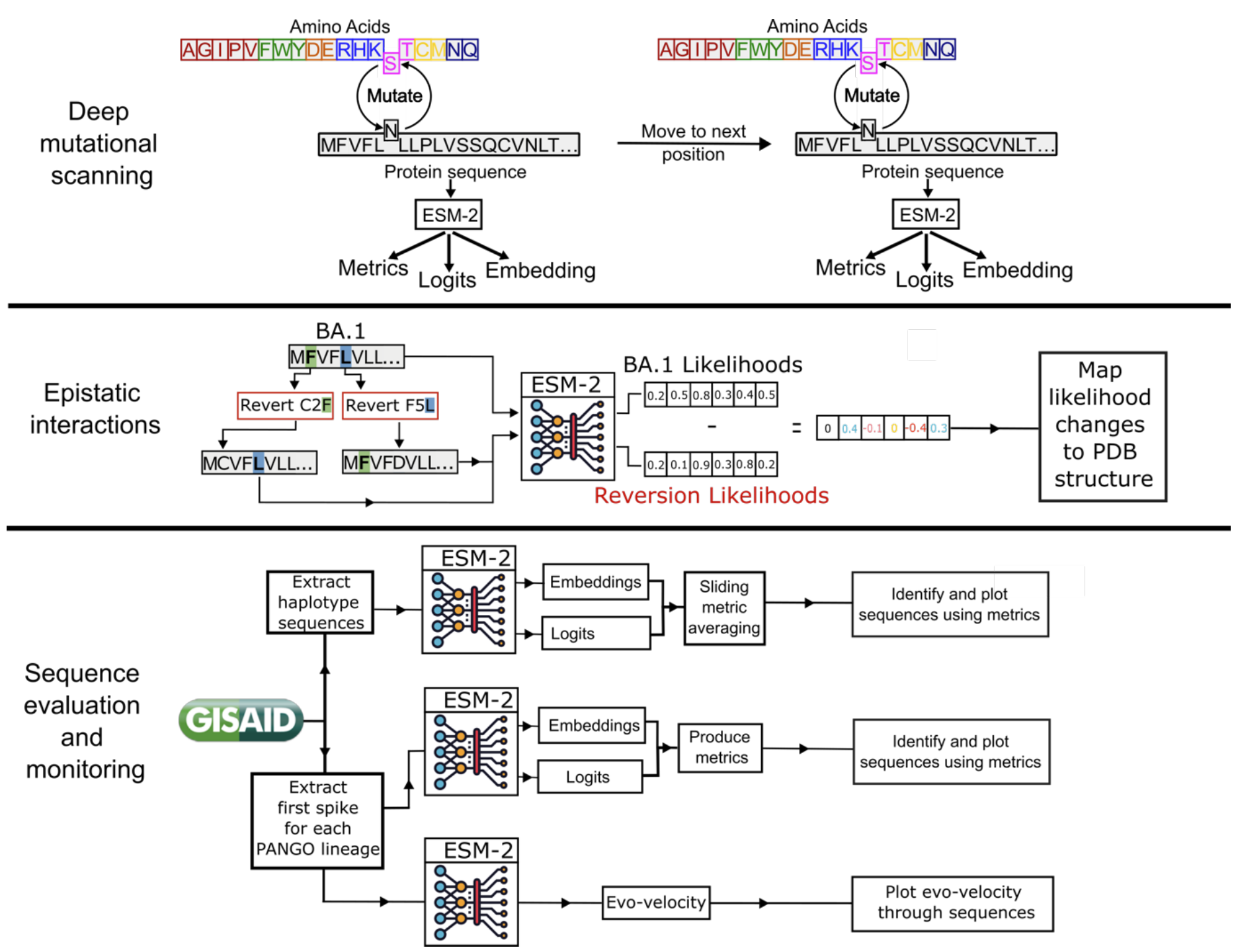
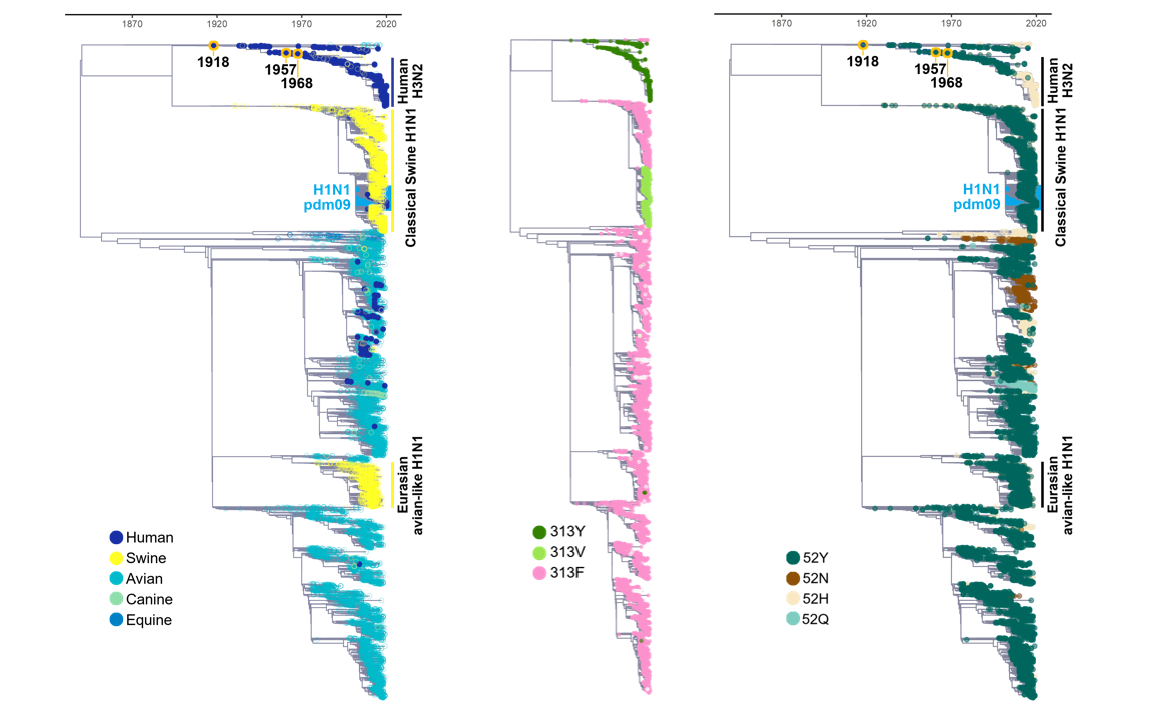
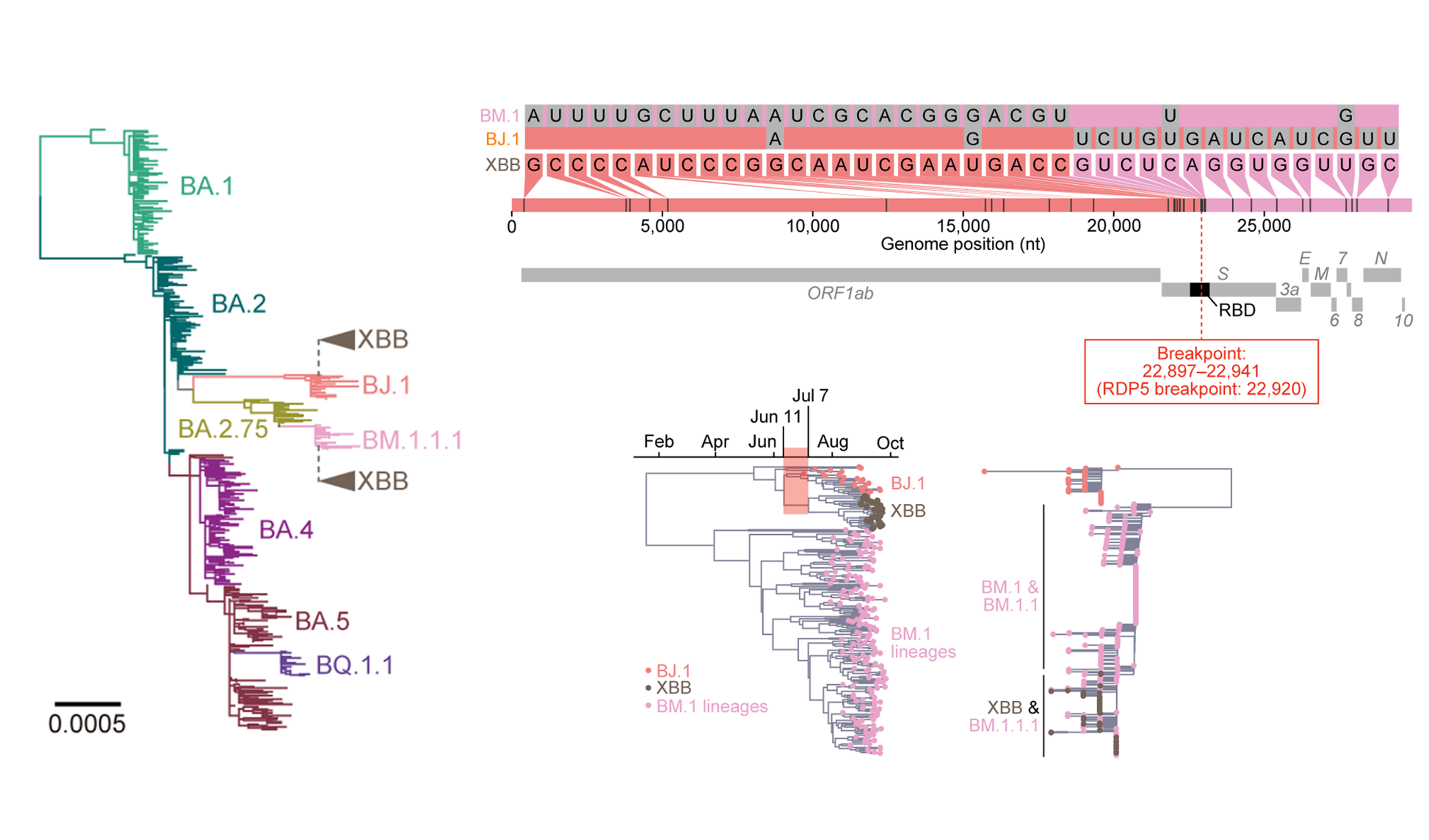
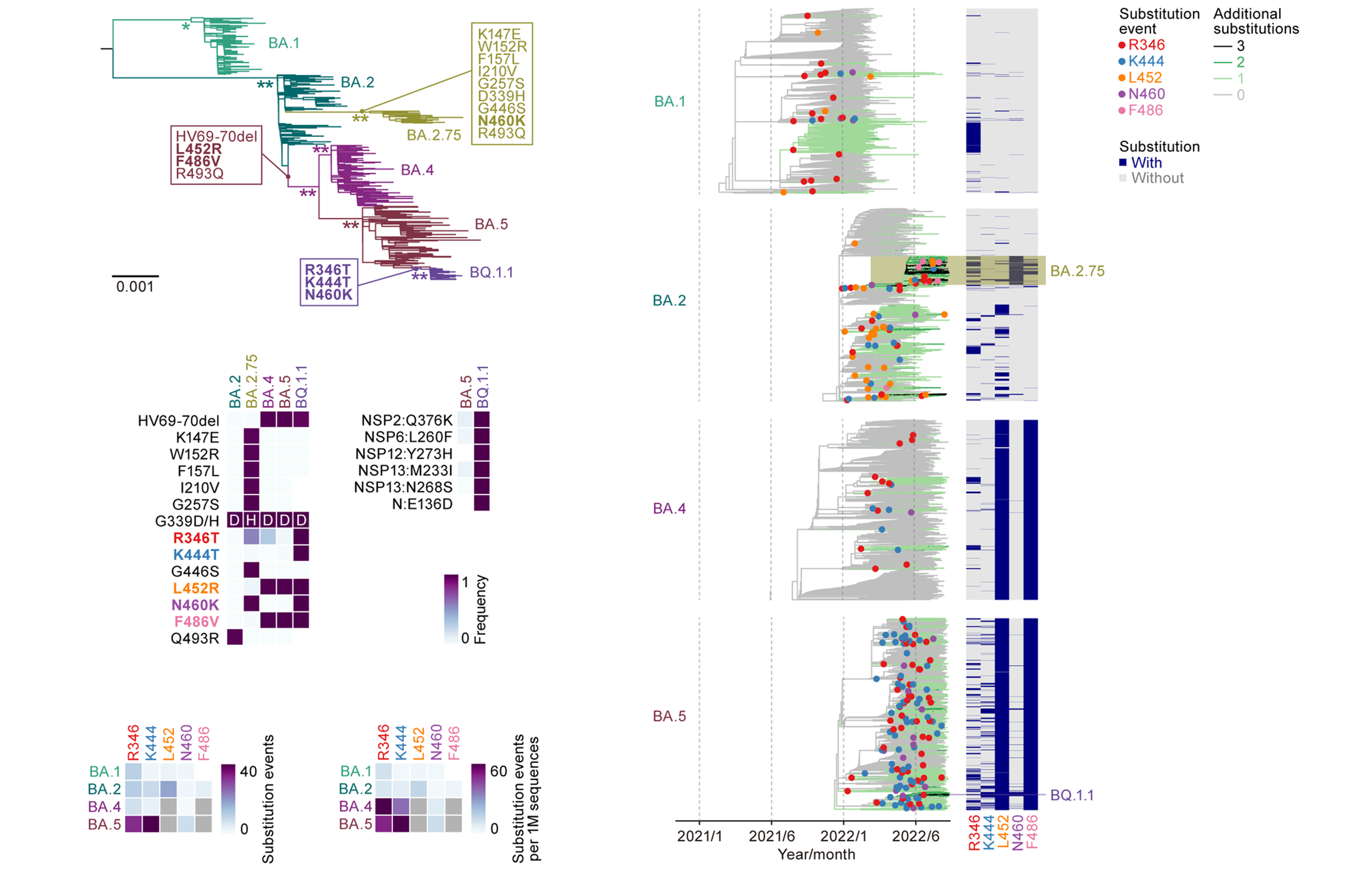
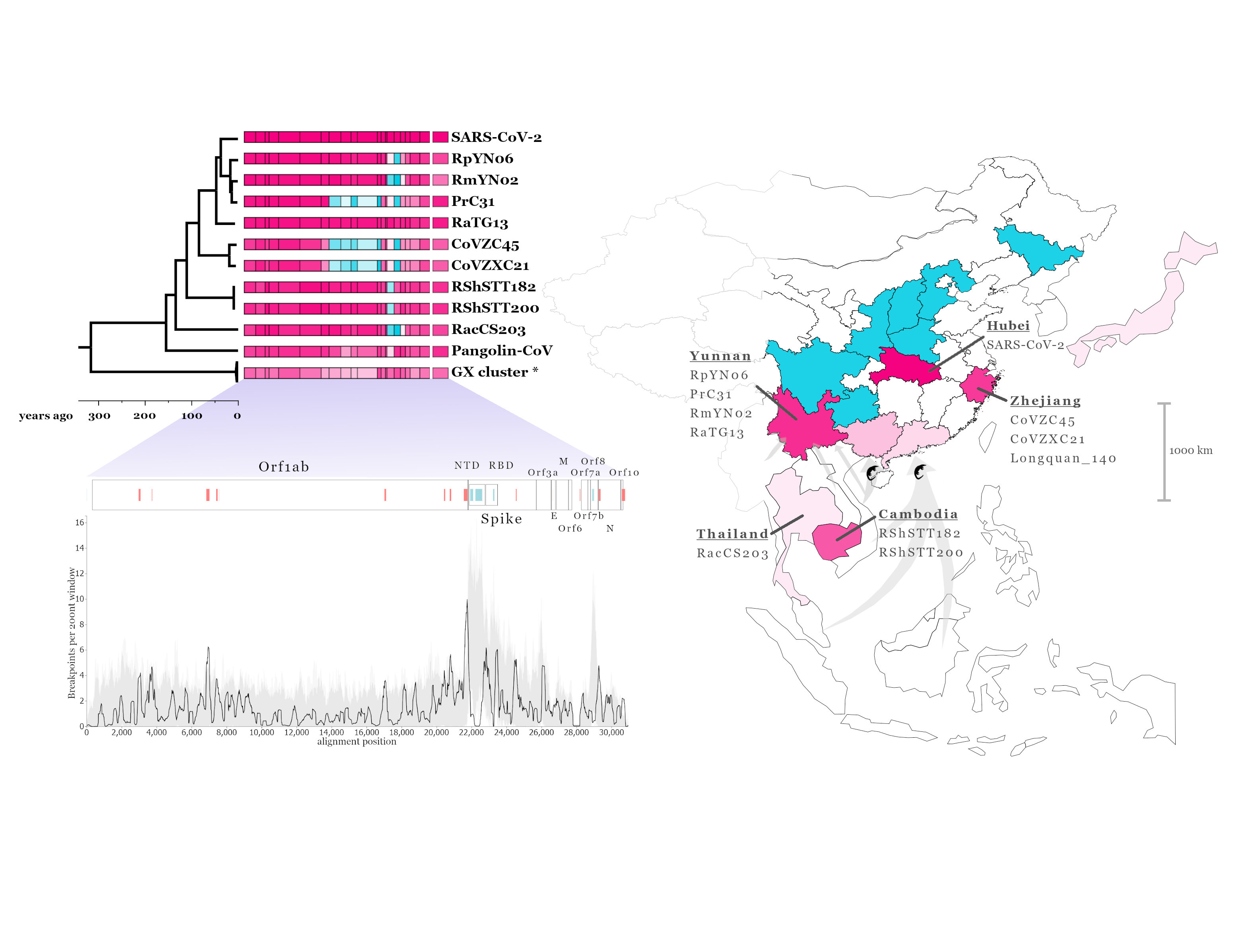
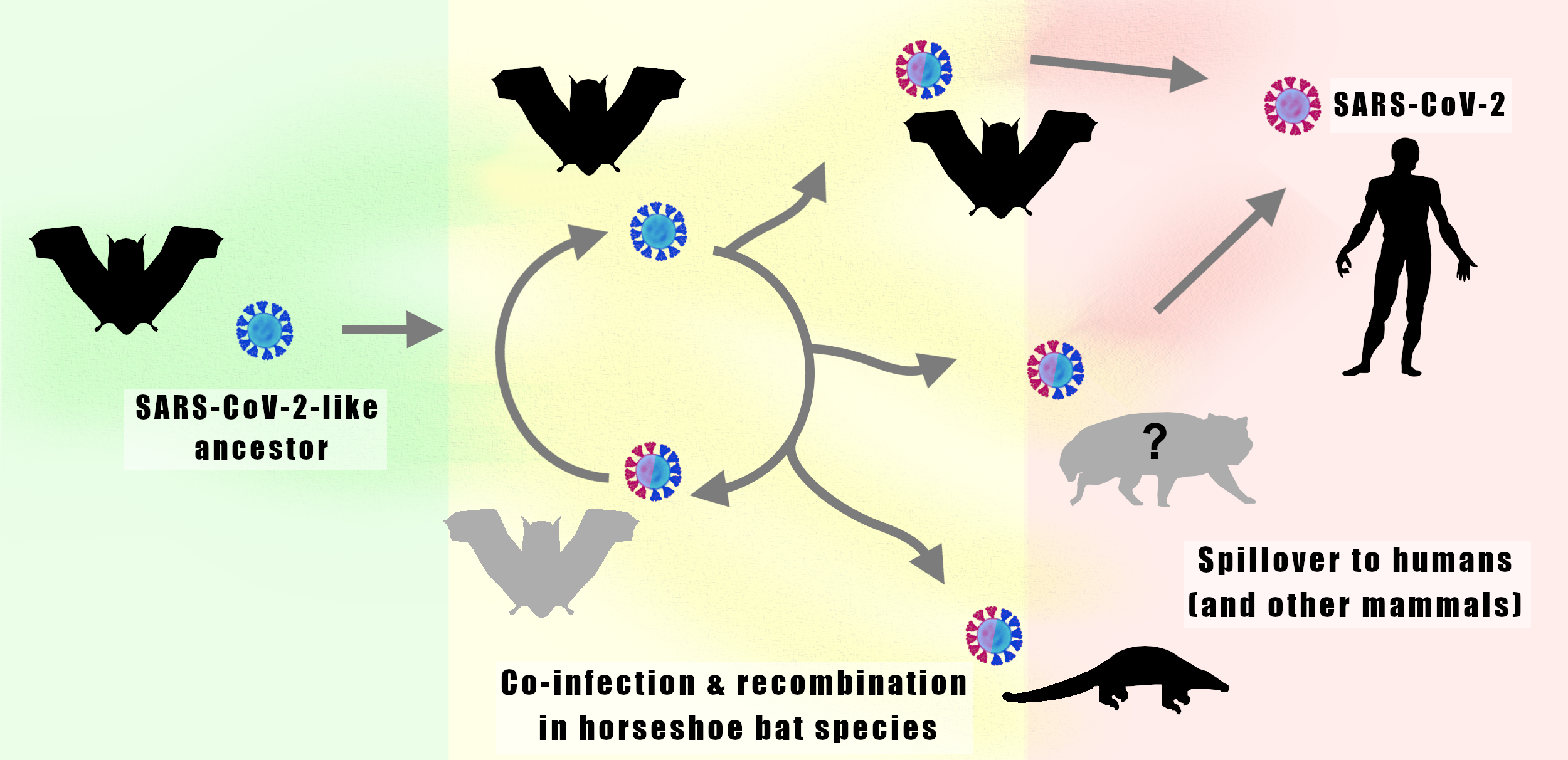
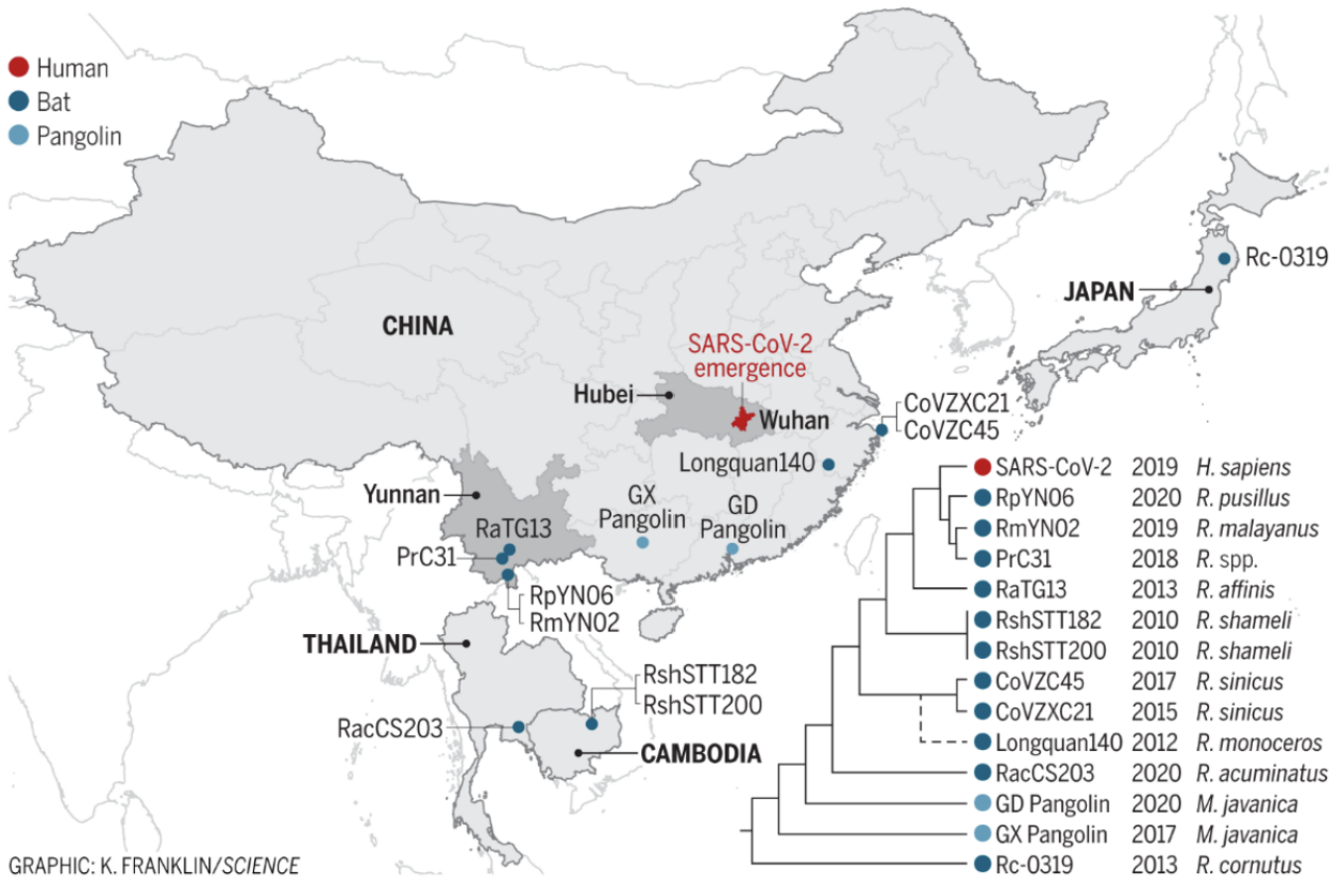
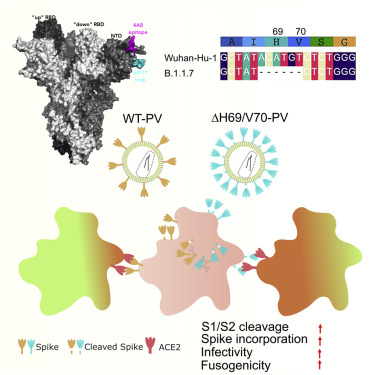
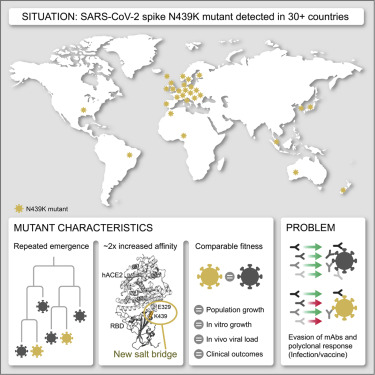
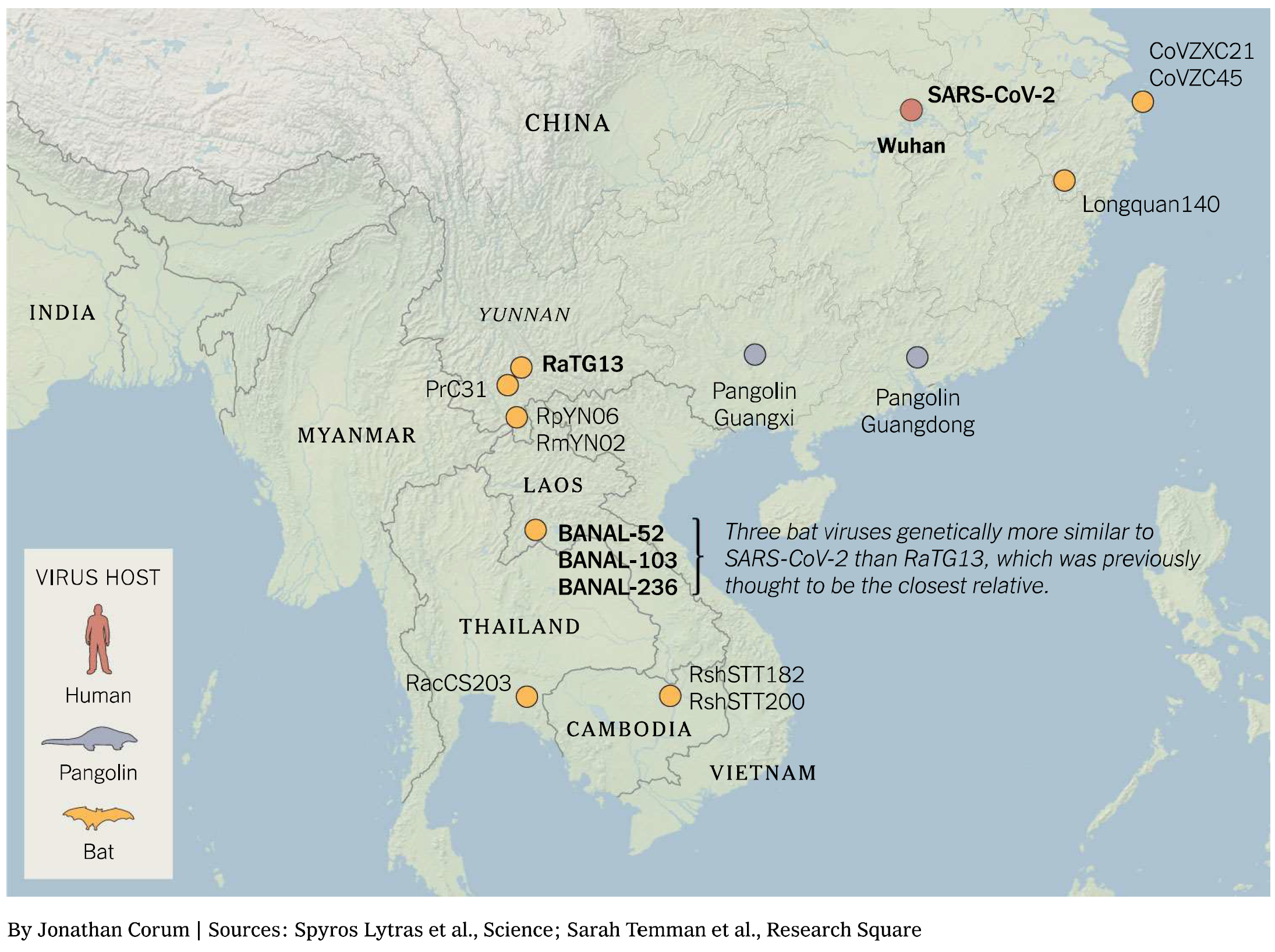
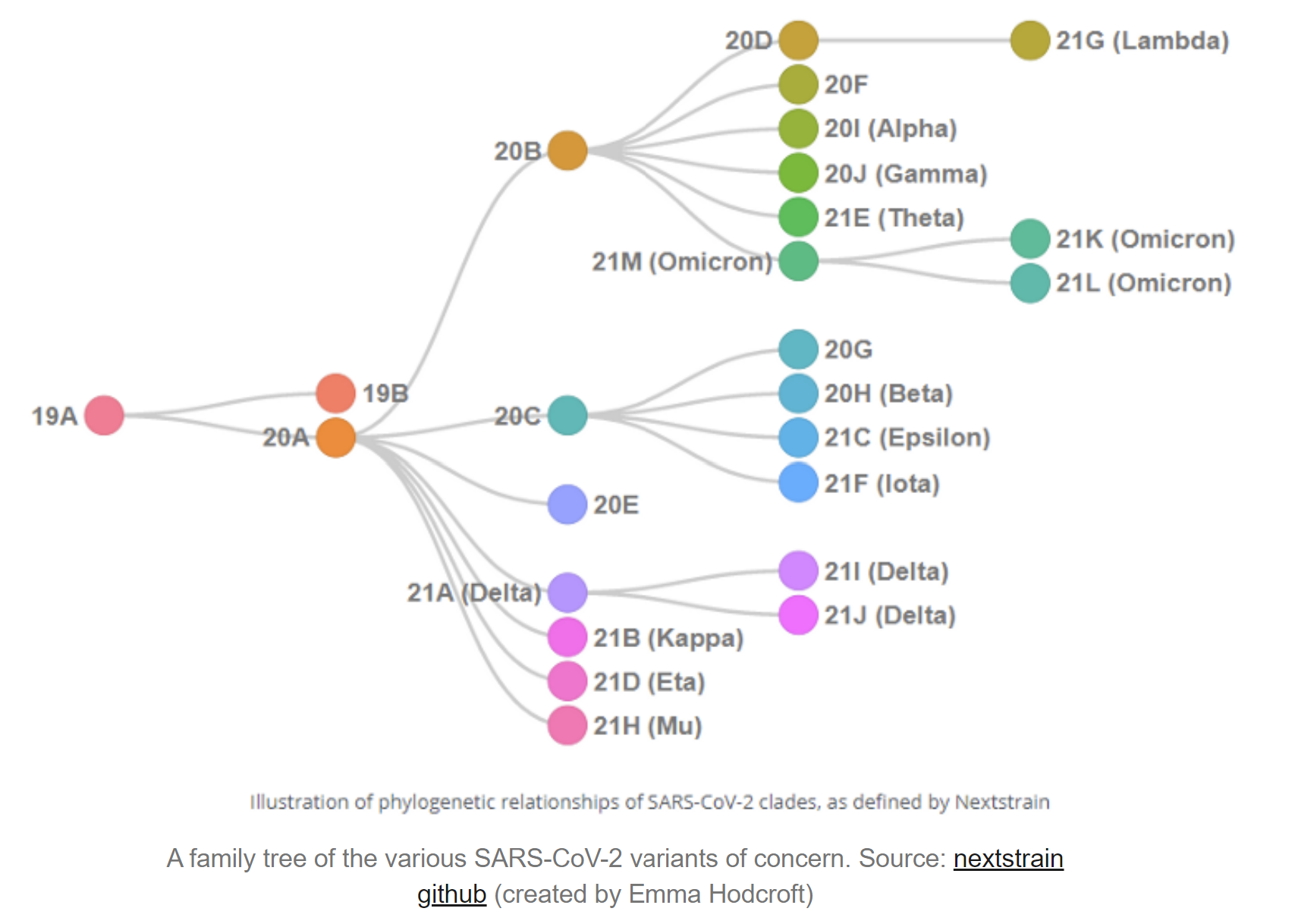
Understanding the origins of the COVID-19 pandemic will help us to stop coronavirus pandemics from happening in the future. Have a watch of my short interview and our animated video explaining how we study the evolutionary history and likely origins of SARS-CoV-2 and its relative coronaviruses.